AI for Cancer Diagnosis: The Success of a Collaboration Led by Mines Paris – PSL
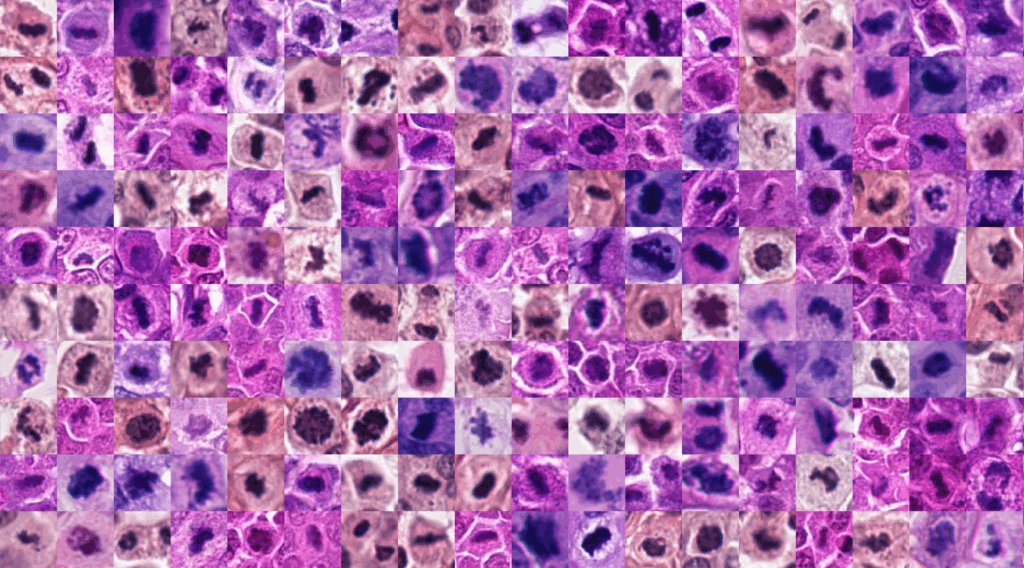

Each year, the prognosis of thousands of cancer patients depends partly on the analysis of mitoses — dividing cells that indicate the aggressiveness of a tumor and can be observed under the microscope on tissue slides. Some mitoses are atypical: their abnormal shapes reveal disrupted cell division processes, often linked to a poorer prognosis.
The problem? Detecting and classifying all these mitoses among hundreds of thousands of cells in a single sample would be practically impossible. That’s why pathologists rely on “hot spots,” small regions of tissue pre-identified for detailed analysis. Even then, the task remains long and difficult — even for seasoned experts.
To tackle this challenge, the Mitosis Domain Generalization Challenge (MIDOG) 2025 was launched. Organized as part of the MICCAI conference — the most prestigious event in medical image analysis — this international competition brought together researchers, engineers, and clinicians around two key objectives (Tasks):
Detect mitoses automatically in varied tissue samples (human and canine tumors, inflammatory zones, etc.);
Classify mitoses as normal or atypical — a crucial step for refining cancer diagnoses.
Beyond its medical importance, the competition specifically aimed to develop robust algorithms capable of performing well regardless of species, tumor type, staining technique, or scanner used. Because of the wide heterogeneity of data across hospitals, robustness remains one of the major challenges of digital pathology.
In artificial intelligence, the concept of domain generalization refers to training methods designed to make algorithms less dependent on the specific features of data from one hospital or imaging device. In practice, this means training models to recognize essential biological structures — even when images come from different sources, stains, or scanners.
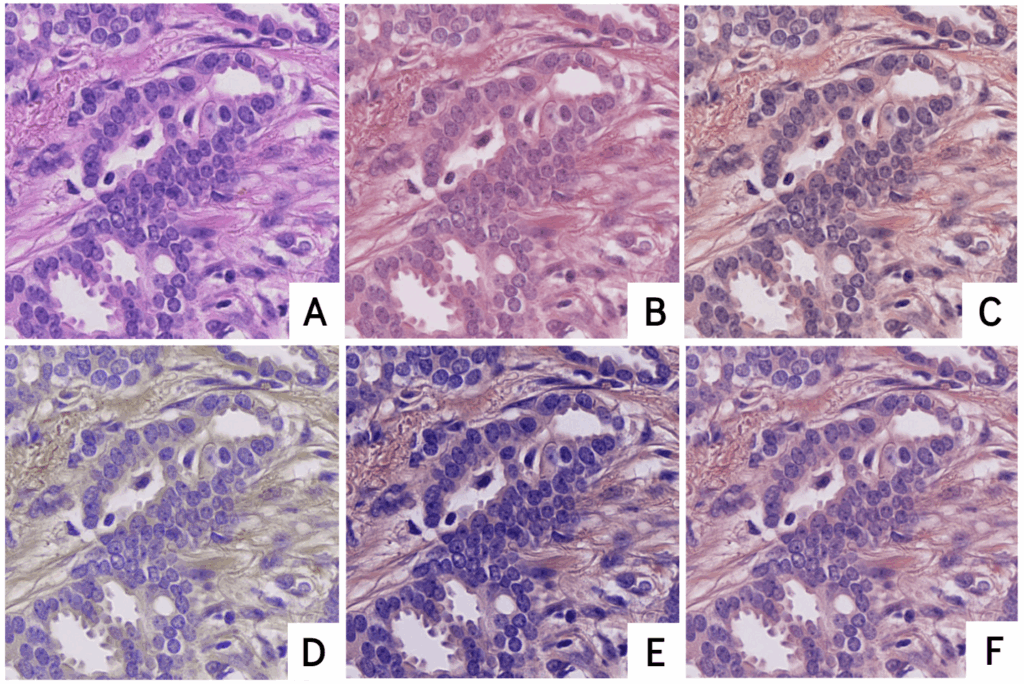
Illustration of the image augmentation process:
(A) Example of a raw microscopic image fragment (640 × 640 pixels) stained with the H&E method, commonly used to differentiate tissue components.
(B–F) Variations of this same image after color normalization processing, simulating the natural differences in staining that may occur from one laboratory to another.
The Mines Paris – PSL team, in collaboration with Institut Curie, CHU de Nantes, and Sanofi, achieved remarkable results: 2nd place for detection (Task 1) and 1st place for atypical mitosis classification (Task 2).
The project was primarily conducted by two PhD students:
Raphaël Bourgade, PhD candidate affiliated with CBIO (Mines Paris – PSL), Institut Curie, and CHU de Nantes, co-supervised by Anne Vincent-Salomon, Delphine Loussouarn, and Thomas Walter.
Guillaume Balezo, PhD candidate affiliated with CBIO (Mines Paris – PSL), CMM (Mines Paris – PSL), and Sanofi, co-supervised by Étienne Decencière, Albert Pla Planas, and Thomas Walter.
They were supported by a strong team effort involving Hana Feki (Master’s student), Lily Monnier (postdoc), Alice Blondel (PhD student), and Matthieu Blons (postdoc).
For Mines Paris – PSL, this dual success at MIDOG 2025 positions its teams at the forefront of artificial intelligence for health. It also highlights the richness of collaborations between academic institutions, hospitals, and industry.
Detecting dividing cells is not just a theoretical challenge — it’s a crucial step in improving cancer treatment. Thanks to AI, Mines Paris – PSL is helping make this process more precise, faster, and more reliable.
Raphaël Bourgade, Guillaume Balezo, Hana Feki, Lily Monier, Matthieu Blons, Alice Blondel, Delphine Loussouarn, Anne Vincent Salomon, and Thomas Walter, Robust Pan-Cancer Mitotic Figure Detection with YOLOv12, oct. 2025.
Guillaume Balezo, Hana Feki, Raphaël Bourgade, Lily Monnier, Matthieu Blons, Alice Blondel, Étienne Decencière, Albert Pla Planas, and Thomas Walter, Efficient Fine-Tuning of DINOv3 Pretrained on Natural Images for Atypical Mitotic Figure Classification in MIDOG 2025, sep. 2025.

AI continues to revolutionize many fields, including medicine. The Santé Numérique seminar of theInstitut des Transformations Numériques (ITN) of Mine...